Image registration is an important preliminary task
in many image processing problems like reconstruction of 3D information from
2D views or motion estimation from temporal sequences. The purpose of image
registration (also called image matching) is to geometrically align one image
(the floating or source image) to another one (the reference or target image)
so that voxels (or pixels in 2D) representing the same underlying structure
may be superimposed. A standard framework for image registration consists
in minimizing a cost function or maximizing a "similarity measure" that
expresses the pixel or voxel similarity of the images to be aligned. Due
to its large variety of sensors, 3D medical imaging is certainly one of
the first application field. Applications range from computer-assisted surgery
to the analysis of sequences of functional images used for example to follow
the evolution of diseases. In fact, it greatly improves both diagnosis and
therapy. The registration may concern images from the same modality or images
from different modalities (multimodal image matching). To compare these images,
many features (edges, surfaces, voxels, etc.) and similarity measures are
now available. Besides, transformations range from rigid transformation,
with a small number of parameters, to deformable image warps, depending
on several thousands or millions of parameters (figure below - T is the
transformation). We consider both rigid and deformable image matching problems.
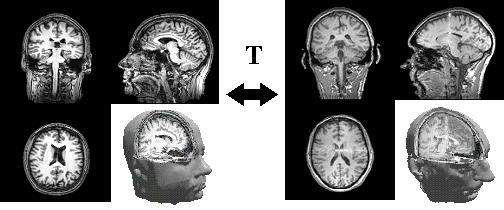
Source image
Target image
Standard cost functions, based on voxel similarity measures,
are highly non-linear, non-convex, exhibit many local local minima and thus
yield hard optimization problems. On the one hand, for rigid matching of single
modal images (Magnetic Resonance Images) we have considered the parallelization
of a general purpose global optimization based on random sampling and evolutionary
principles: the differential evolution algorithm. The inherent parallelism
of evolutionary algorithms is used to devise a data-parallel implementation
of differential evolution. The data-parallel algorithm can yield subvoxel
accuracy and exhibits an almost linear speedup. On the other hand, for deformable
inter-subject matching of 3D MR images (the registration of two 256^3 MRI
typically involves the optimization of 200,000 parameters, and takes several
hours on a standard workstation), we have proposed a general fully parallel
approach based on the simulation of stochastic differential equations. This
approach is naturally suited to massively parallel implementations, yielding
high quality registrations and excellent relative speedups. To conclude, this
work has shown the relevance of the parallelization for medical image registration
and the suitability of the data-parallel programming model.